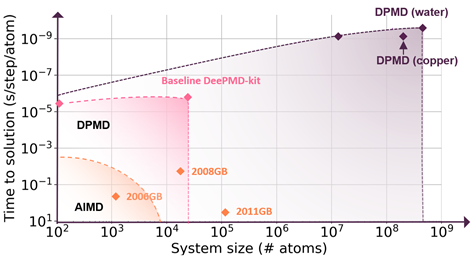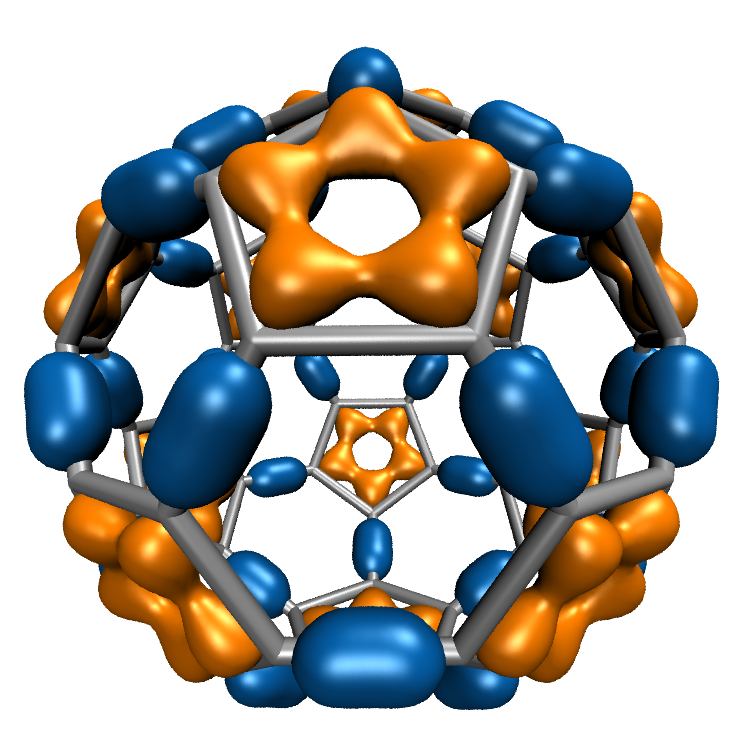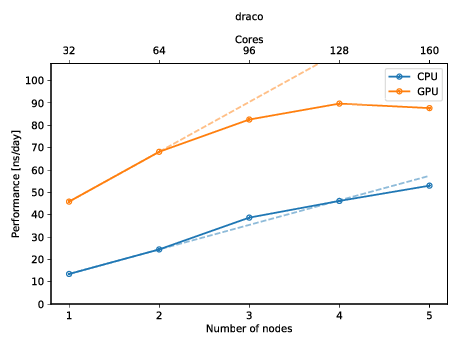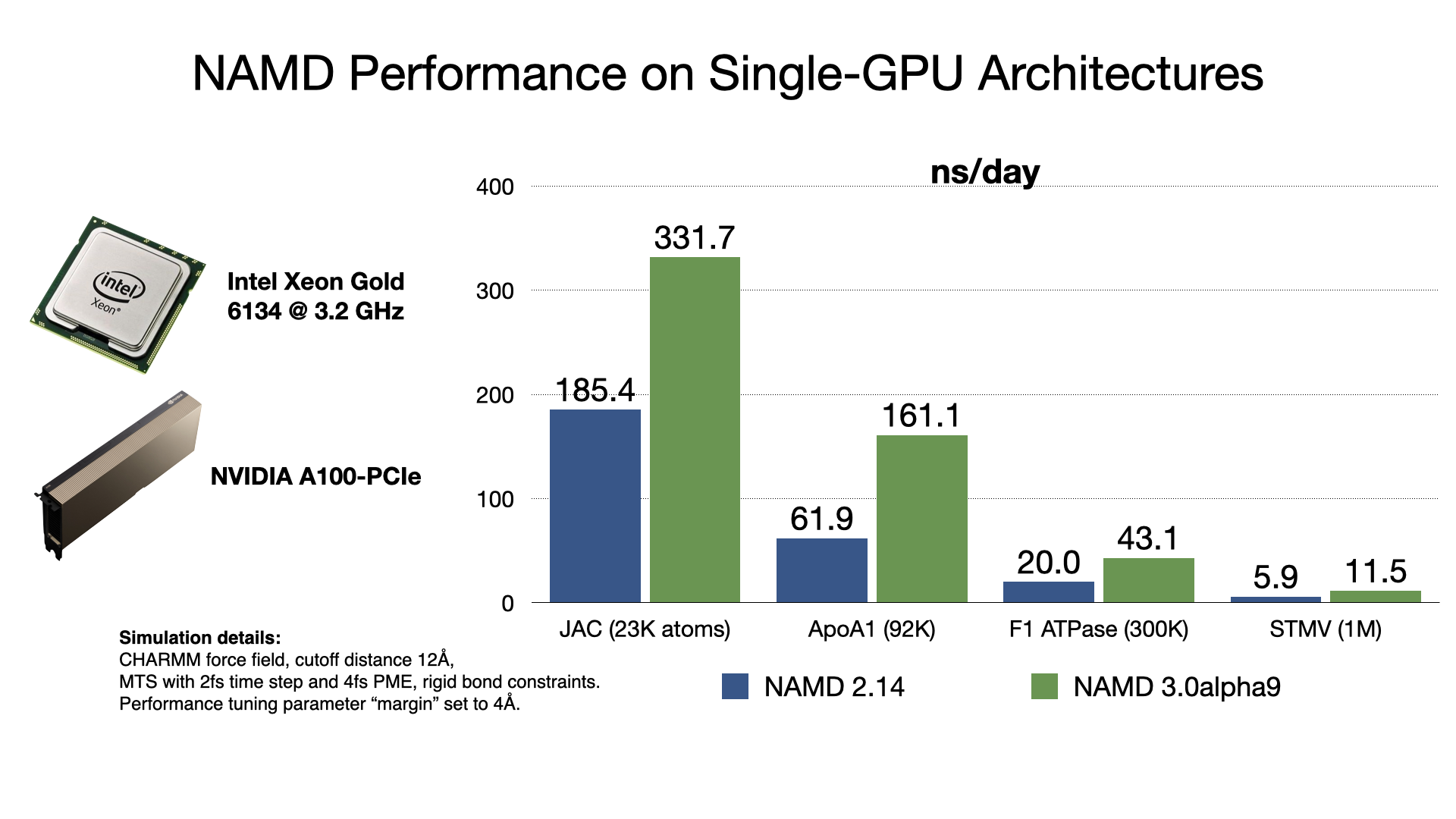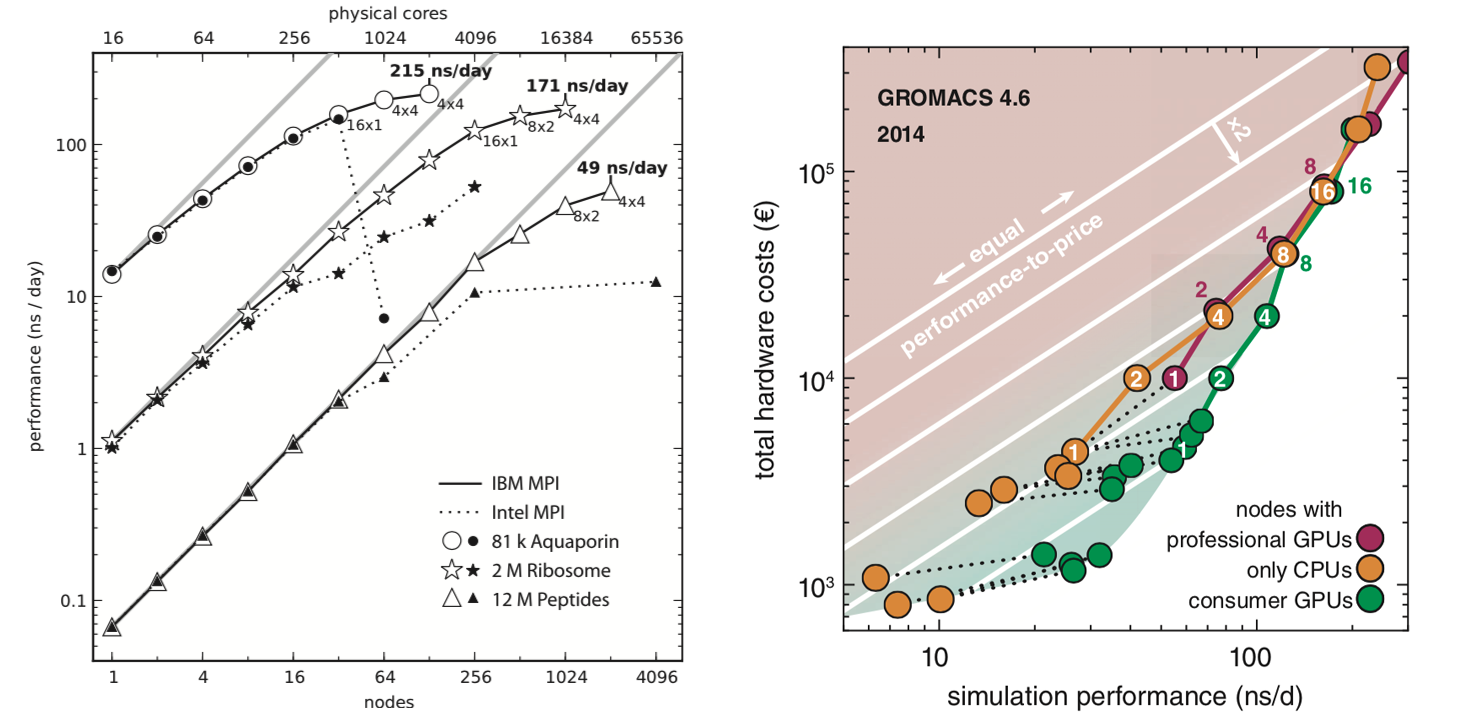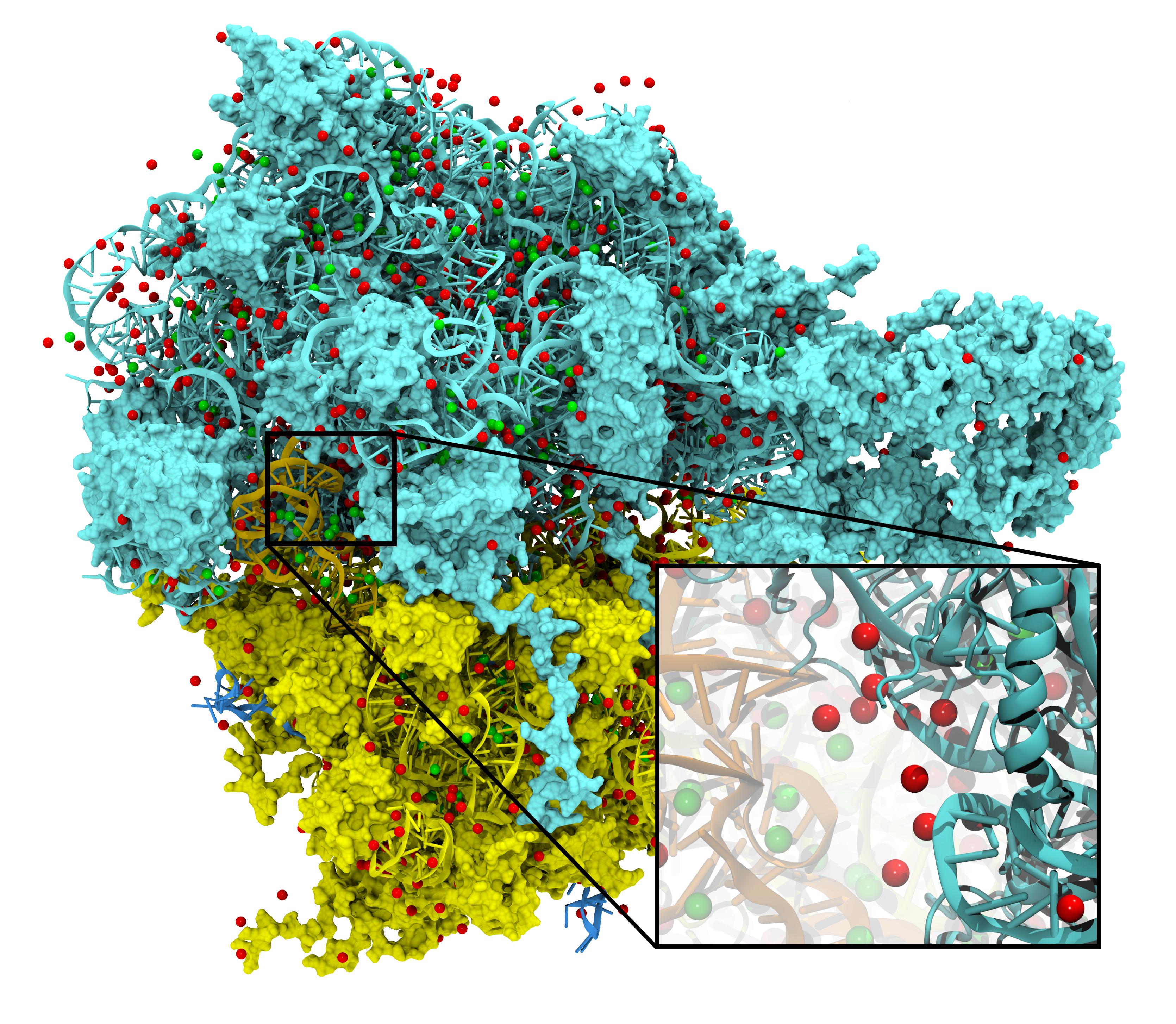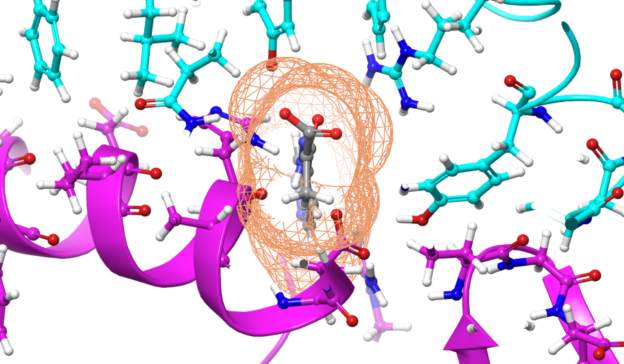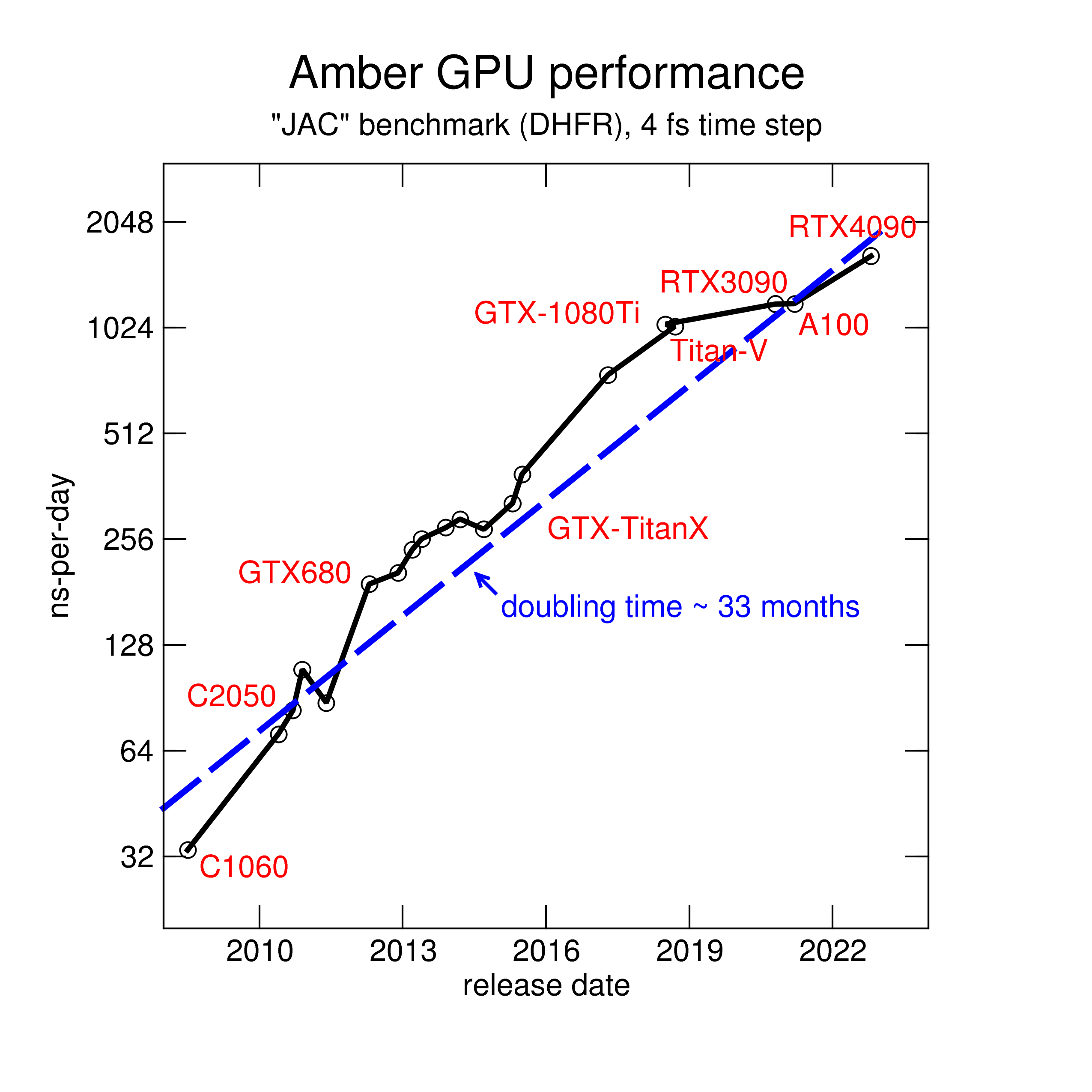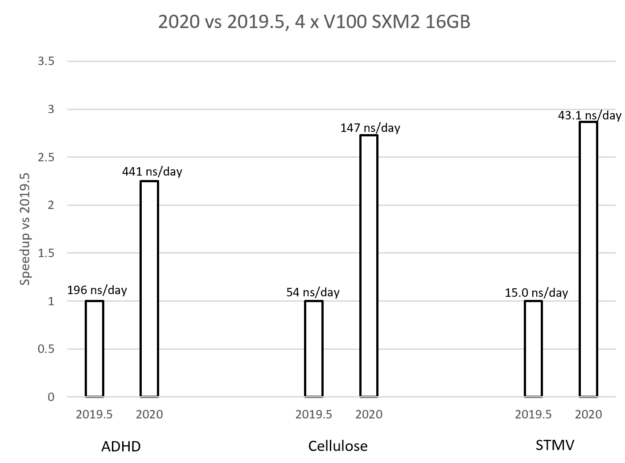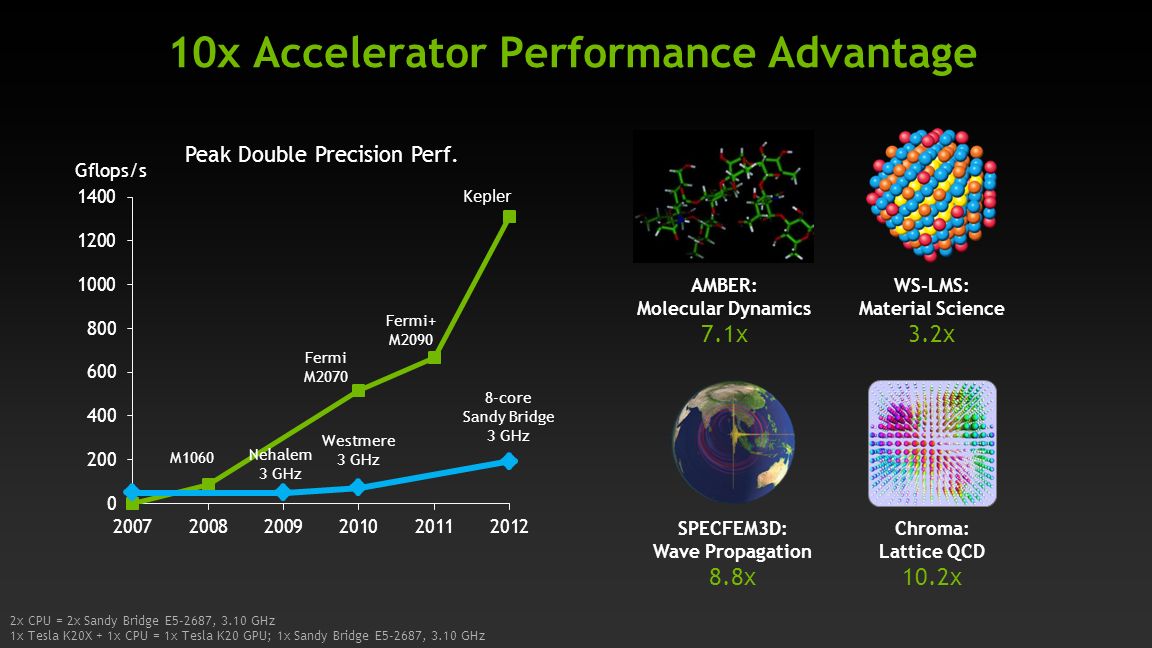
August Accelerated Computing CPU Optimized for Serial Tasks GPU Accelerator Optimized for Parallel Tasks 10x Performance 5x Energy Efficiency. - ppt download

Heterogeneous CPU+GPU-Enabled Simulations for DFTB Molecular Dynamics of Large Chemical and Biological Systems | Journal of Chemical Theory and Computation
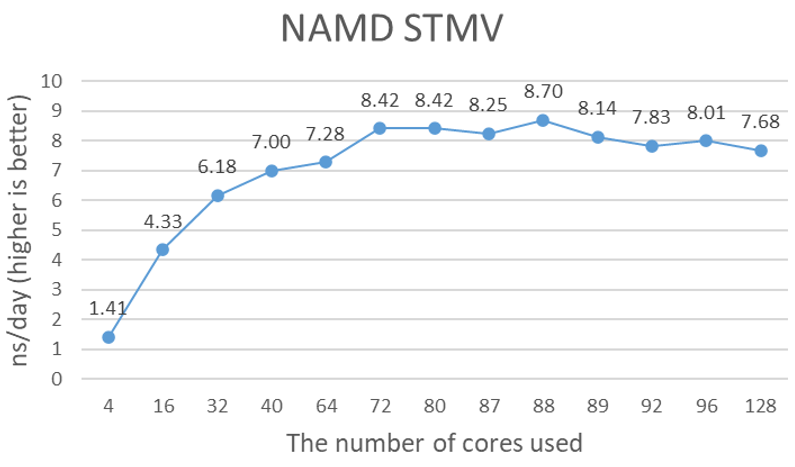
Molecular Dynamics Simulations with Dell EMC PowerEdge XE8545 Server and NVIDIA A100 | Dell Technologies Info Hub

A GPU-Accelerated Machine Learning Framework for Molecular Simulation: HOOMD-blue with TensorFlow | Theoretical and Computational Chemistry | ChemRxiv | Cambridge Open Engage
PLOS ONE: GPU-Accelerated Molecular Dynamics Simulation to Study Liquid Crystal Phase Transition Using Coarse-Grained Gay-Berne Anisotropic Potential