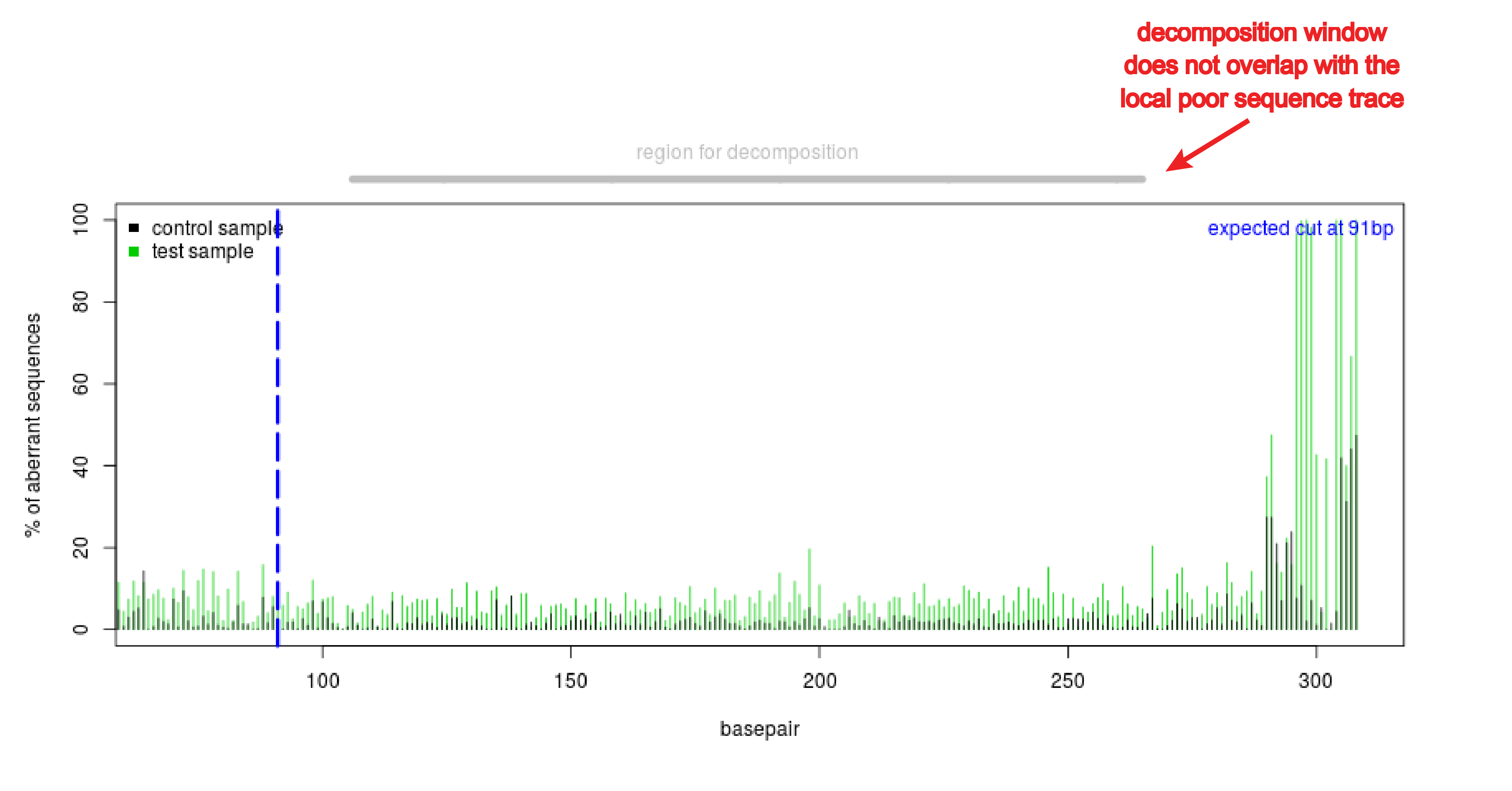
Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases - ScienceDirect
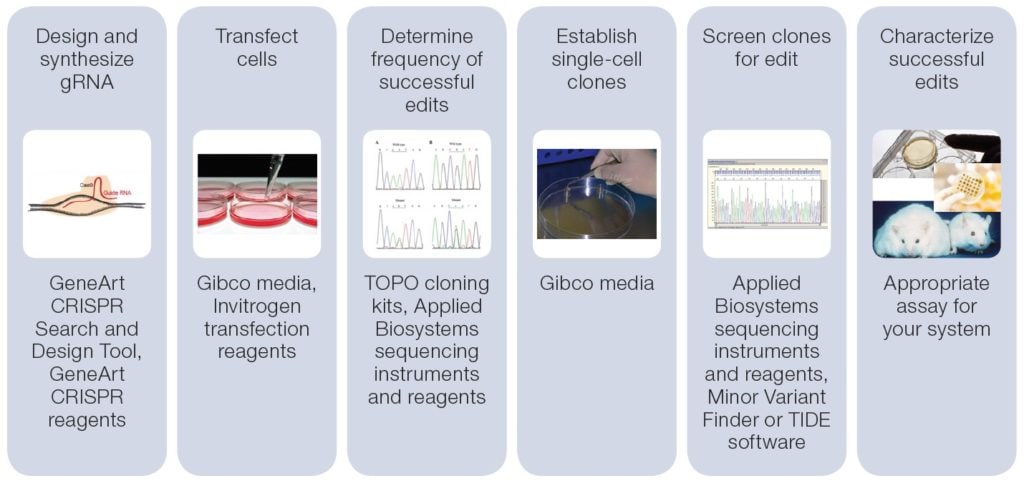
CRISPR-Cas9 Genome Editing Guide – Finessing the technique and breaking new ground - Behind the Bench
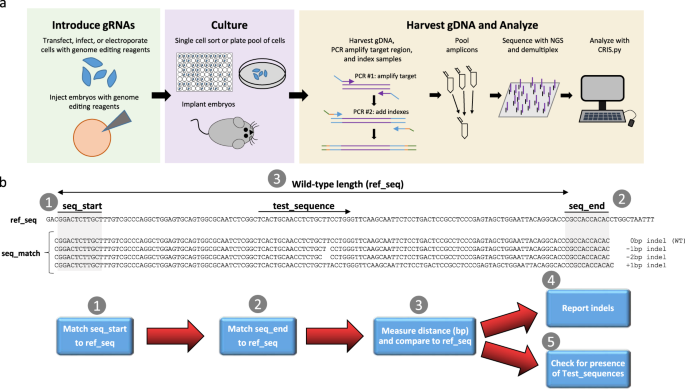
CRIS.py: A Versatile and High-throughput Analysis Program for CRISPR-based Genome Editing | Scientific Reports

TIDE sequencing analysis of light-mediated indel mutation HEK293T cells... | Download Scientific Diagram

CRISPR/Cas9-Directed Reassignment of the GATA1 Initiation Codon in K562 Cells to Recapitulate AML in Down Syndrome: Molecular Therapy - Nucleic Acids
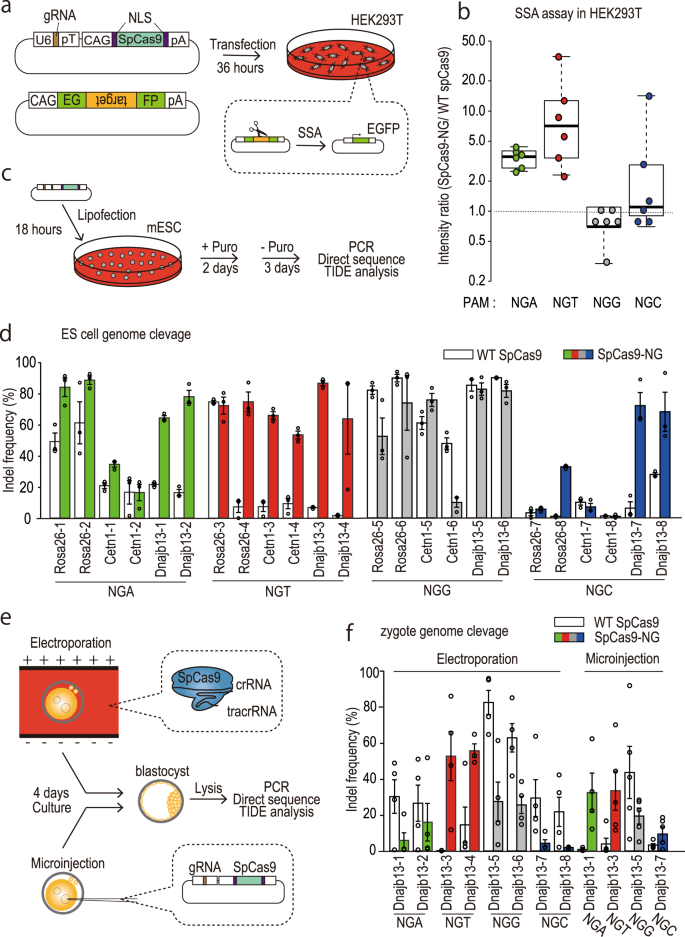
Precise CAG repeat contraction in a Huntington's Disease mouse model is enabled by gene editing with SpCas9-NG | Communications Biology
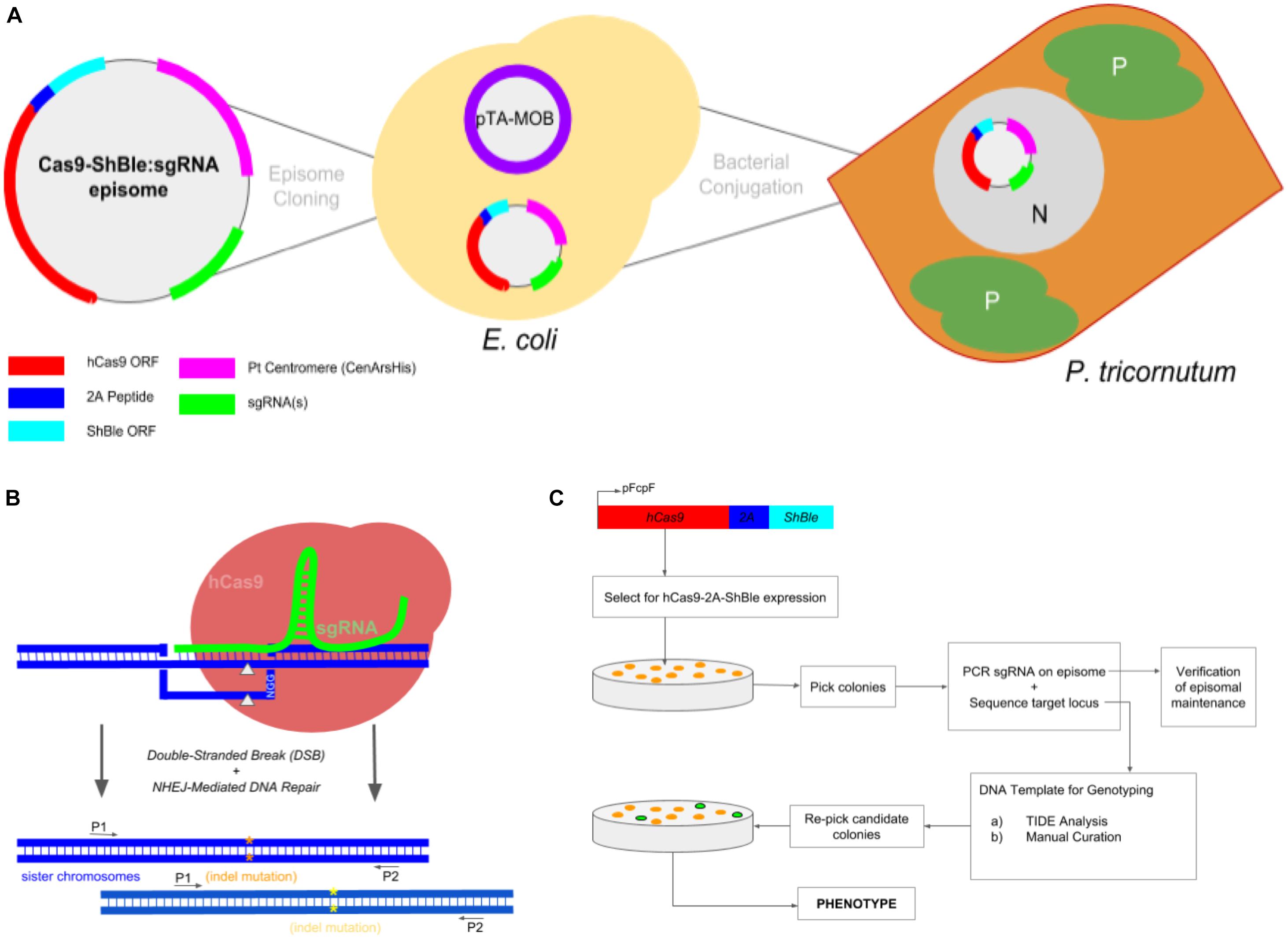
Frontiers | Multiplexed Knockouts in the Model Diatom Phaeodactylum by Episomal Delivery of a Selectable Cas9 | Microbiology

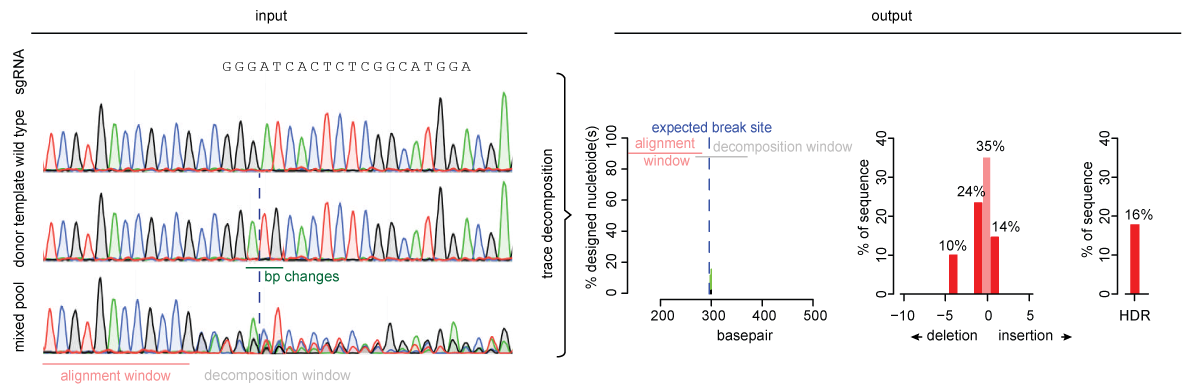




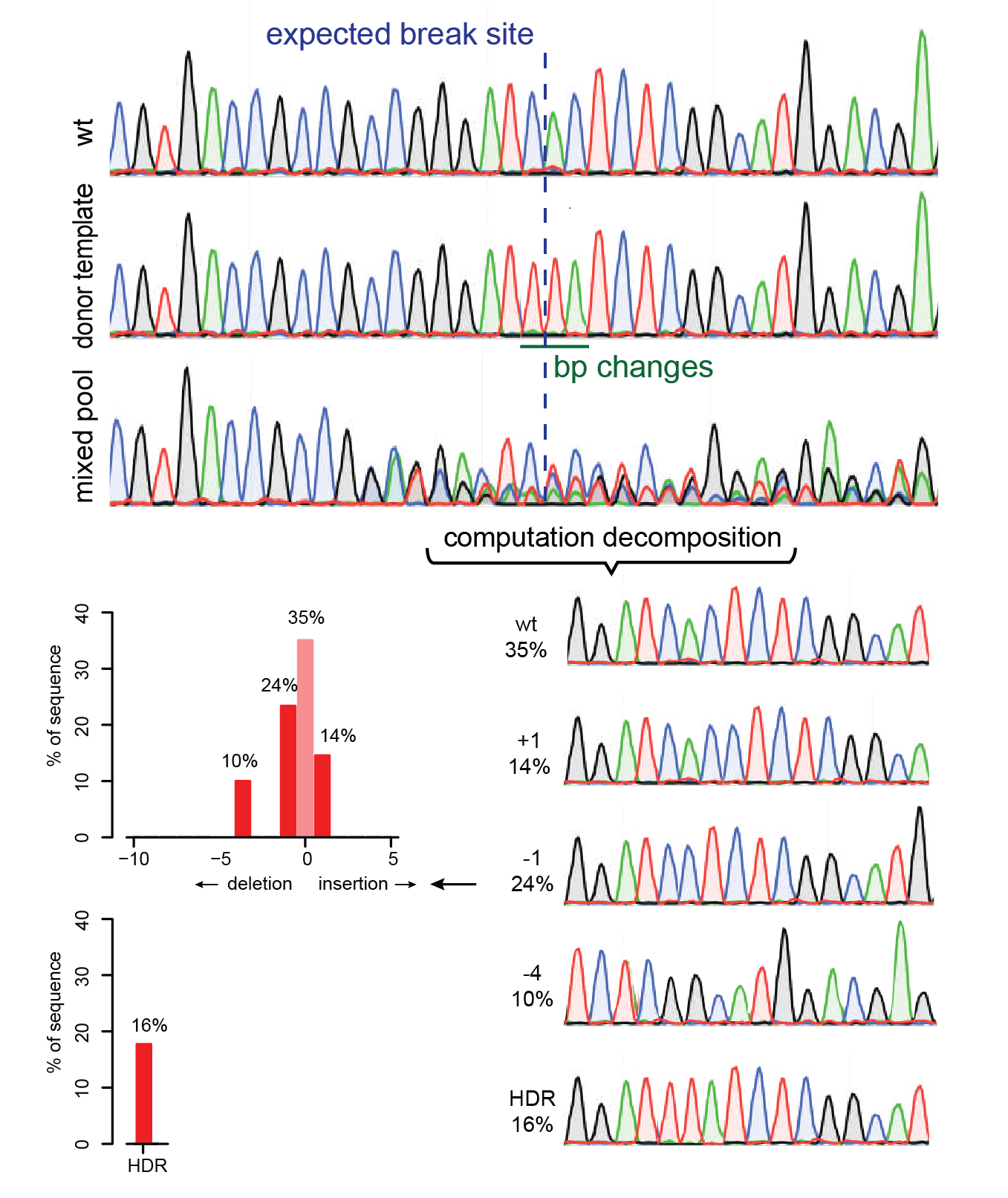

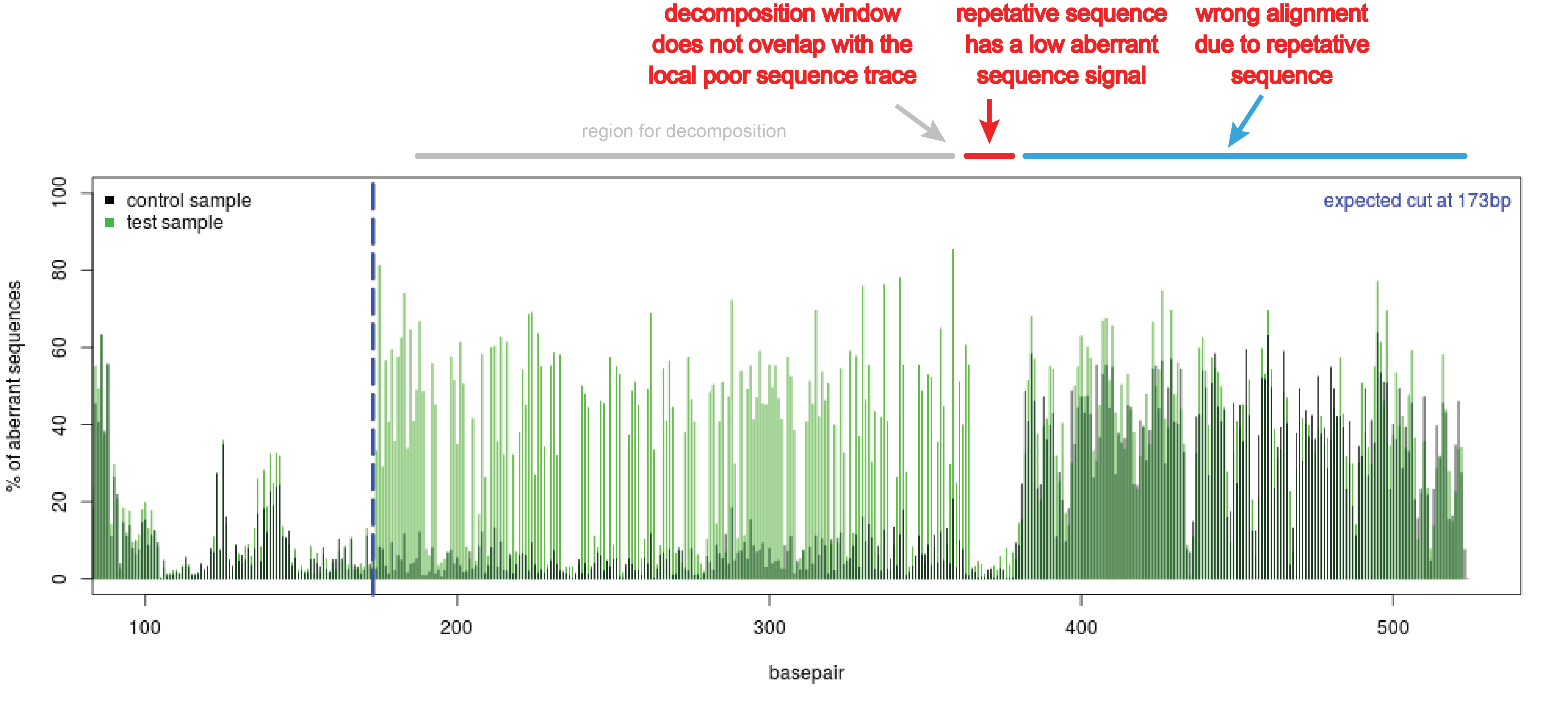

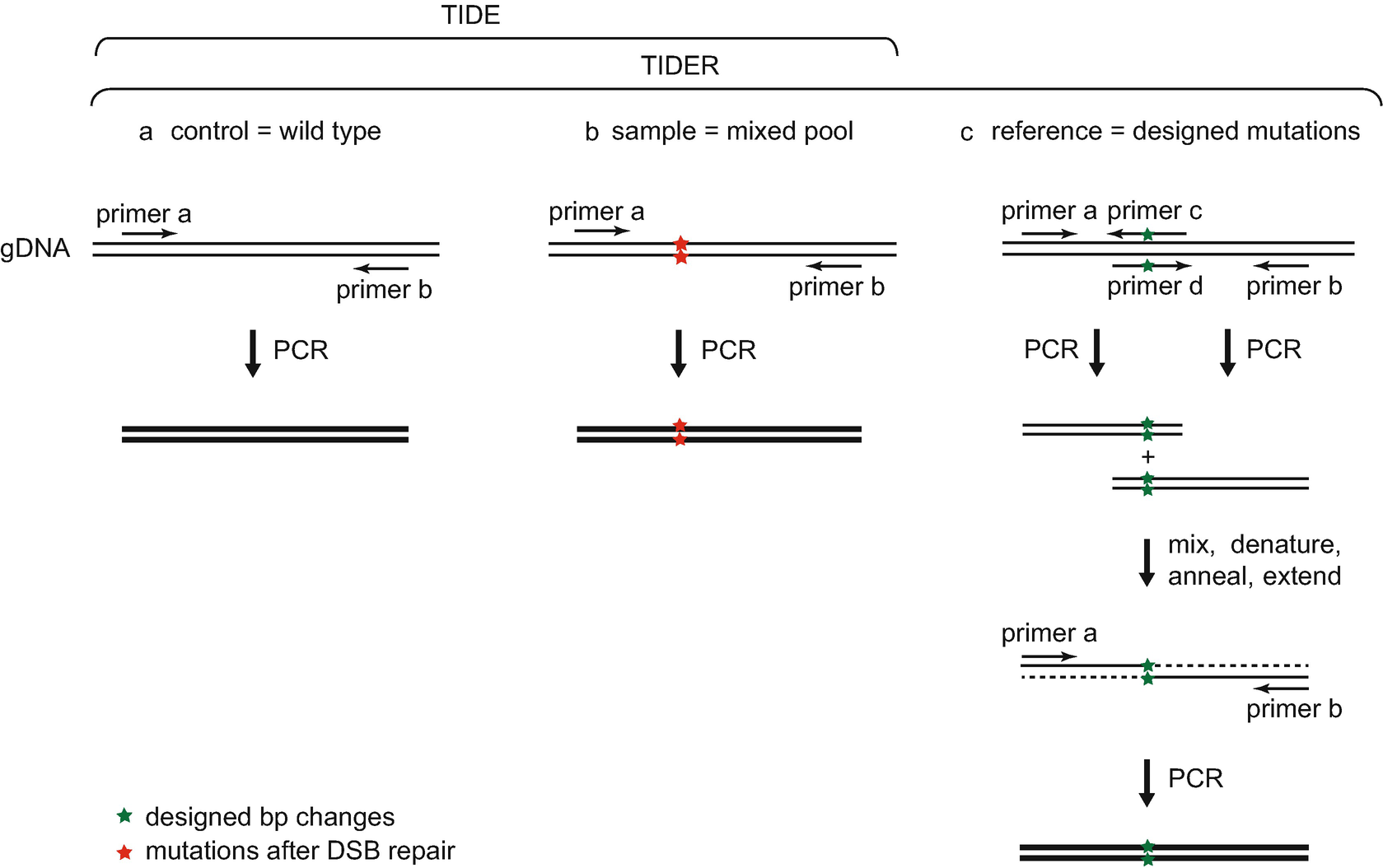
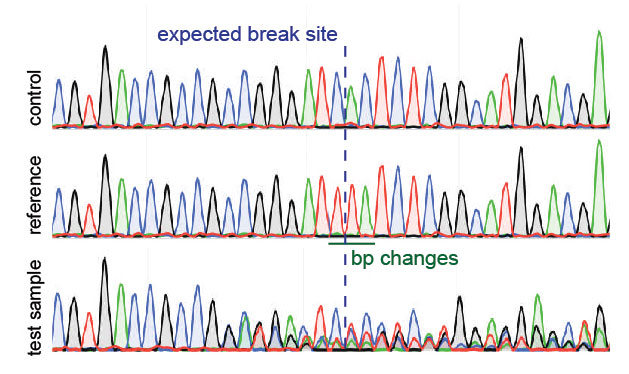
![A dual sgRNA approach for functional genomics in Arabidopsis thaliana [OPEN] | bioRxiv A dual sgRNA approach for functional genomics in Arabidopsis thaliana [OPEN] | bioRxiv](https://www.biorxiv.org/content/biorxiv/early/2017/08/21/172676/F1.large.jpg)